Public hearing for data scientist disciplined for "posting information that went against the narrative the employer was promoting, which is that COVID-19 vaccines are safe and effective” is now set.
18-20 FEB 2026: come to observe - in person or remotely. The outcome of this hearing will signal what kind of Public Service—and what kind of Canada—we are moving toward.
THIS PAGE IS REGULARLY UPDATED IN PREPARATION FOR MY FPSLREB HEARING. ALL INFORMATION ABOUT THE HEARING AND HOW TO OBSERVE IT REMOTELY IS POSTED ON IVIM.ca/hearing.
Updates:
FEB 23: The Exhibits (redacted) from Days 1-3 of the Hearing are now posted at www.ivim.ca/hearing/book-of-documents. The Caselaw (Book of Authorities) submitted by Employer and Grievor will be also made available at www.ivim.ca/hearing/book-of-authorities.
FEB 20: The Hearing Adjourned — Continuation Expected in Summer. The Grievor asks those following the hearing to remain respectful in their comments on social networks.
Several videos, which were previously removed from public viewing as per employer’s instructions, are now open again for public. This includes a Short Film made in 2022 for International Film Festival called “A data scientist's quest for truth":
FEB 15: A new series Seeing Through the Data — The Second Act (A saga of a public servant seeking the truth and the ways to defend it) has been launched to provide more details about my story.
FEB 13: Book of Documents (Evidence of over 200 pages) has been uploaded to FPSLREB portal. The public version of it is also shared with on this Substack. A program code in Python was written to automate the process and can be shared with anyone who needs to organize their documents for the court and any other hearing:
FEB 06: Presentation at Freedom Rising providing the Backgrounder for the Hearing posted:
JAN 30: Instructions on how to attend remotely are posted in the portal (you’ll need to provide me your name and email by MON 16 FEB, and I’ll pass it to FPSLREB director)
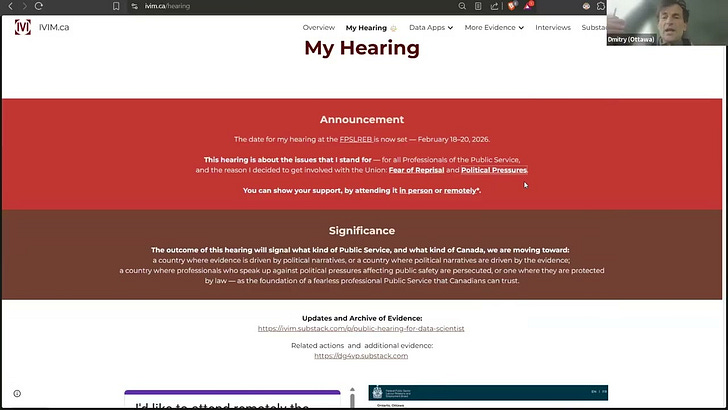
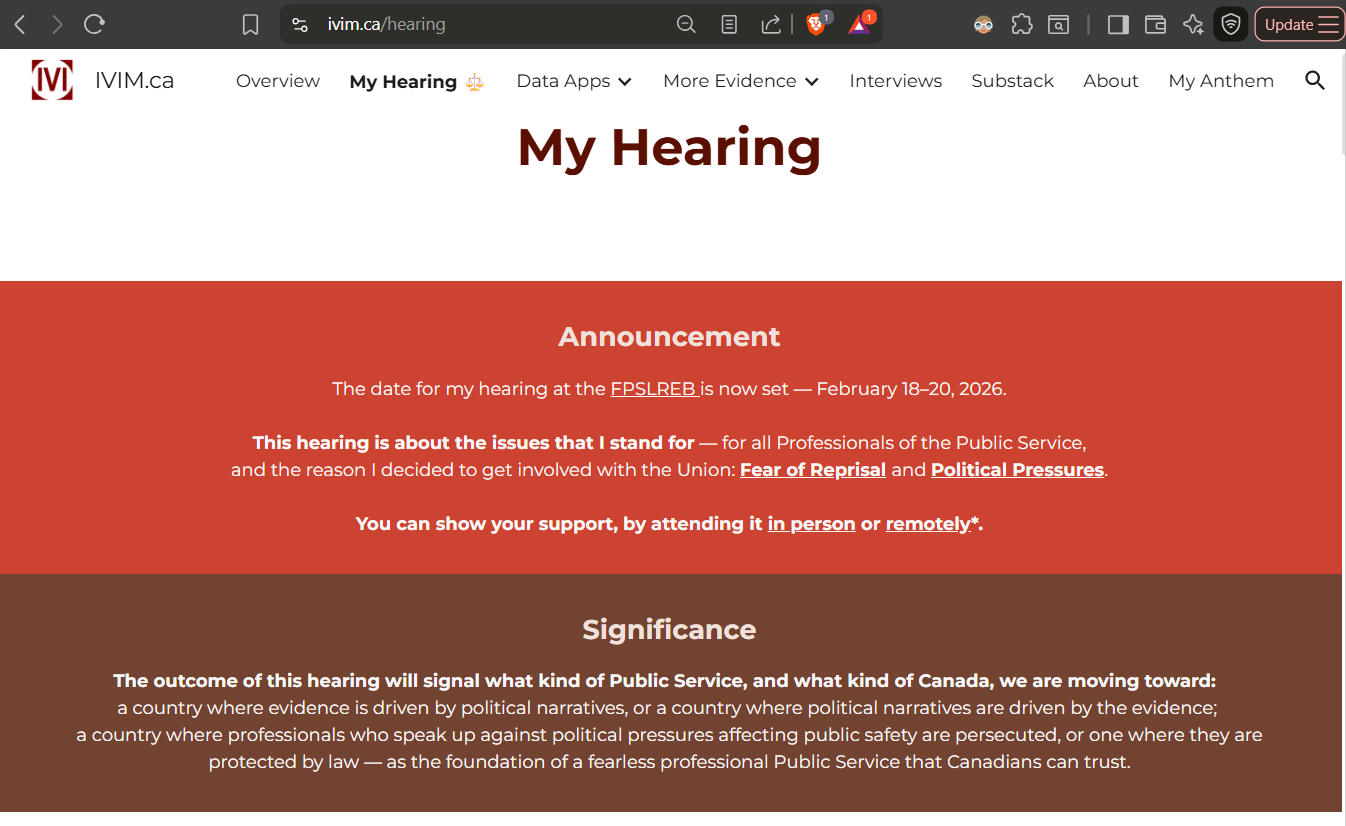
JAN 26: A page is created on IVIM.ca portal dedicated to My Hearing: IVIM.ca/hearing. This is where all details about the Hearing and all related evidence - including my analysis of PHAC and StatsCan data, and communications with my Employer and the Union - will be posted. You may wish to bookmark this page, as this hearing will decide the future of Public Service and Canada.
JAN 19: The dates for My Hearing are announced on both of my portals (en.gorodnichy.ca and www.IVIM.ca) and substacks (IVIM & DG4VP)
15 FEB 2026:
New Series (Novel) in works:
Seeing Through the Data — The Second Act.
(Alternative title: The Data Scientist Who Learned the Truth).
A saga of a public servant seeking the truth and the ways to defend it
DRAFTS Published:
26 JAN 2026: Page is created on IVIM.ca portal
19 JAN 2026: Announcement
Dear Friends,
As just posted on my official website: en.gorodnichy.ca/, the date for my hearing at the FPSLREB is now set — February 18–20, 2026.
Please mark your calendar and come to observe it - in person or via videoconference (link soon to be provided), to be a witness of this decisive moment in the Canadian Public Service history.
I’ve been waiting for this for hearing for more than three years…
In October 2022, my employer suspended me for five days without pay for discussing official data related to COVID-19 vaccines in Government of Canada data science forums (video) and elsewhere, in particular at the Freedom Convoy (video). I am accused that I “willingly posted information that is contrary to the message that the Government of Canada is promoting which is that COVID-19 vaccines were safe and effective”. I am saying that the data showed what it showed, I just made them easier to understand, using my professional data science skills, for the benefit of all public servants and all Canadians.
The outcome of this hearing will signal what kind of Public Service—and what kind of Canada—we are moving toward.
A dedicated page is created on my portal (www.ivim.ca/hearing) to provide more details about this case and all related evidence and background.
Below are the first two pieces of evidence - the video recordings of two incidents where I’m alleged of misconduct (presentation at the lunch-and-learn data science seminar at my work and my speech at Freedom Convoy on 4th February 2022), followed by another video recording (where I call Chief Science Officer to request addressing concerning post-marketing vaccine data), which I plan to use to ask if this was also misconduct
More is coming. Stay tuned. It is worth revisiting what was exactly happening back then - to mark the fourth year anniversary of one of the biggest peaceful demonstrations in the history of humankind.
Evidence used against the Grievor:
2022-02-04: “One Year since Vaccination. What we have learnt - using Open Canada Data & Data Science” - Lunch-and-Learn Seminar, MS Teams (30 mins)
Slides (Source: https://open-canada.github.io/vitals/OneYearOfVaccineInCanada-deck-meetup-2022-02-04.pdf
2022-02-04: Senior Data Scientist addresses Freedom Convoy protesters in Ottawa (9 mins)
Background and Transcript:
Snapshots from IVIM.ca (2022/03/29)
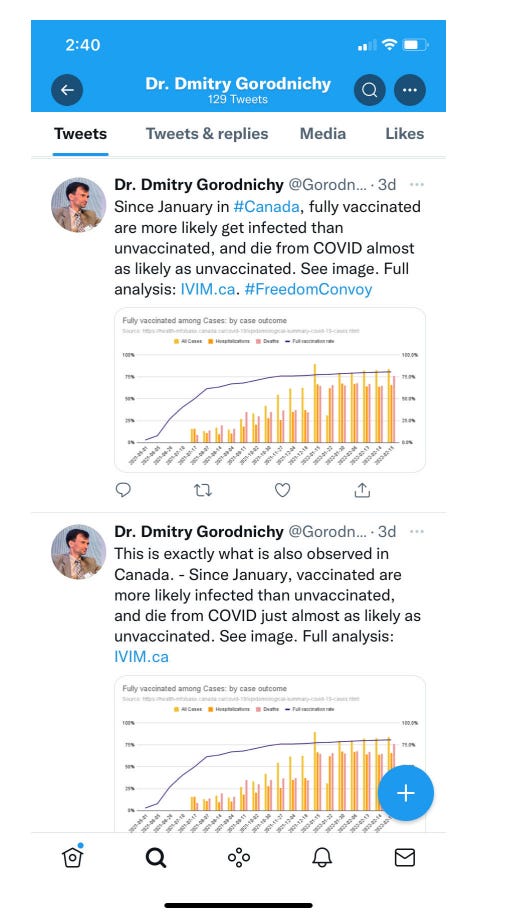
Snapshot from Twitter:
Evidence provided by the Grievor
2022-05-13: Follow up with Chief Science Advisor - Re: Report on COVID-19 Vaccine Efficacy & Safety (4 mins)
You can read my Open Letter to Chief Science Advisor of Canada from this article:
Grievor’s presentations and evidence:
Open-Canada GitHub pages:
PHAC data:
Evidence used by Employer:
Snapshot of www.ivim.ca (2022-03-29)
Snapshot from Twitter:
White Paper (February):
Presentation at Data Science Lunch-and-Learn seminar (Slides)



!['Seeing Through the Data — The Second Act'. Preface to a New Series: Setting the Stage. [DRAFT]](https://substackcdn.com/image/youtube/w_728,c_limit/x8RanqovIVc)





How about writing a short summary? I could not even figure out which department you worked for. It will make a difference whether it's CBSA ....or Dept of Fisheries. It is a quandary. If one works for a company, it is not good form to criticise that company except internally. One is expected to resign, lose salary, and then criticise. Hmmm. Maybe we have to argue that the vaccine mandates were so harmful, that normal standards do not apply. Unfortunately, lawyers and plaintiffs are saying the courts are refusing to listen to medical arguments, saying it is not their job.
Anybody who believes the fake fax was "safe and effective" with all the contrary scientific and medical proof is a Pathological Liar that believes their own lies (and those lies of other liars). All clinics in NA have published a timeline that PL's become medically insane after only a few months of believing their own lies. The barking dumb dogs they call the CanGov is a perfect example of insane liars!